Summer course in analysis of high throughput data for population genetics 2025
General info
Registration is closed except for UCPH PhD students
Date: August 4-8, 2025
Place: Copenhagen, Denmark
Organized by: The Department of Biology and GLOBE, University of Copenhagen
Price:
- On site: Free for all PhD students at Danish, Faroese and Greenlandic universities. 200 Euro for all other students.
- Virtual: Free
Includes:
- On site: All teaching. Food and accommodation are NOT included in the course fee.
- Virtual: includes steaming, server access, and online teaching assistants.
Sign up: Registration for UCPH PhD students
Contact: For questions write to cphsummercourse@gmail.com
Credit and diploma: The course is 2.5 ETCS credits and you will recieve a diploma.
Africa BioGenome Project participants: The course will be offered as part of the African BioGenome Project Open Institute East and Central Africa Regional Workshop.
github with code
Content
The course is a comprehensive introduction to a number of topics and common research tools used in analyses of next-generation sequencing (NGS) data. Topics include: genetic drift, natural selection, population structure, recent and ancient admixture, f-statistics and demographic inference. We will cover the theoretical concepts but the main focus is practical use of the methods. Lectures will be combined with hands-on computer exercises.
Intended Learning Outcome
After the course the student should be able to:
- Use population genetic theory to infer basic population genetics characteristics from genetic data
- Be able to infer ancestry and population structure based on genetic data
- Use NGS data including low depth for population genetic inference.
- Select the optimal strategy for selection scans depending on the population genetics characteristics of the sample population, including taking population structure into account
- Interpret and discuss the results of own analyses and results in the scientific literature
Instructors
Jazmín Ramos-Madrigal

Jazmín Ramos Madrigal is an evolutionary genomicist and Assistant Professor at the University of Copenhagen’s Centre for Evolutionary Hologenomics. Her research combines ancient DNA, genomics, and microbiome studies to explore the evolutionary history of domesticated plants and animals, including maize, grapes, dogs, and wolves. website
Katia Bougiouri

My current research focuses on imputation and local ancesty inference. website
Shyam Gopalakrishnan

Population and statistical genetics website
Martin Sikora

Human and pathogen evolution. The Sikora group works at the intersection of ancient genomics, population genetics and paleoepidemiology and investigates the evolutionary history of humans and our associated pathogens. website
Ida Moltke

The research in my research group is focused on developing and applying statistical and computational methods to genomic data to solve problems in both population and medical genetics. Within population genetics we are mainly interested in trying to gain insights into how the human species has evolved and how the world was peopled. Within medical genetics we are mainly interested in identifying genetic variants that play role in a number of different diseases and traits, such as type 2 diabetes. website
Fernando Racimo

We are interested in using population and quantitative genetics to understand past evolutionary processes, with a particular focus on human evolution and ancient DNA. Our projects include developing tests to detect patterns of ancient selection, creating methods to integrate functional and population genomic data, and inferring demographic parameters using present-day and archaic human genomes. website
Anders Albrechtsen

My group develops statistical and computational methods for analysis of genomic data both for population genetics and medical genomics website
Time and place
The course will take place from Monday August 4 to Friday August 8th 2025 at Biocenteret, Ole Maaloes Vej 5, 2200 Copenhagen N. All lectures and computer exercises will take place in room 4.0.24 (i.e. building 4, ground floor, room number 24). This includes research talks. You can enter the building from two sides as shown shown on this map.
Laptop
You should bring a laptop to the course. We will log into a remote server from the laptop so any laptop will do regardless of operating system.
Course material
The lecture will be based on a large amount of reading material (articles/notes) that should be read in advance - you can find them here once they are finalized (you will get an email with password). The slides used during the lectures will be made available right before the lectures.
Program (Preliminary)
Monday - Introduction to population genetics and NGS data
09:00 - 09:15 Welcome (Ida Moltke)
09:15 - 10:15 Lecture 1: Intro to basic population genetic terms and concepts (Fernando Racimo)
10:30 - 12:00 Computer exercises I
12:00 - 01:00 Lunch (on your own)
01:00 - 01:45 Lecture 2: Intro to basic NGS data, data processing and formats (Anders Albrechtsen)
01:45 - 03:15 Computer exercises II (break at 02.15)
03:30 - 04:15 Research talk: Understanding genetic architecture of disease in Greenland using population genetics by Frederik Stæger.
04:15 - 07:00 Reception/social mixer
Tuesday - Analysis of NGS data and imputation
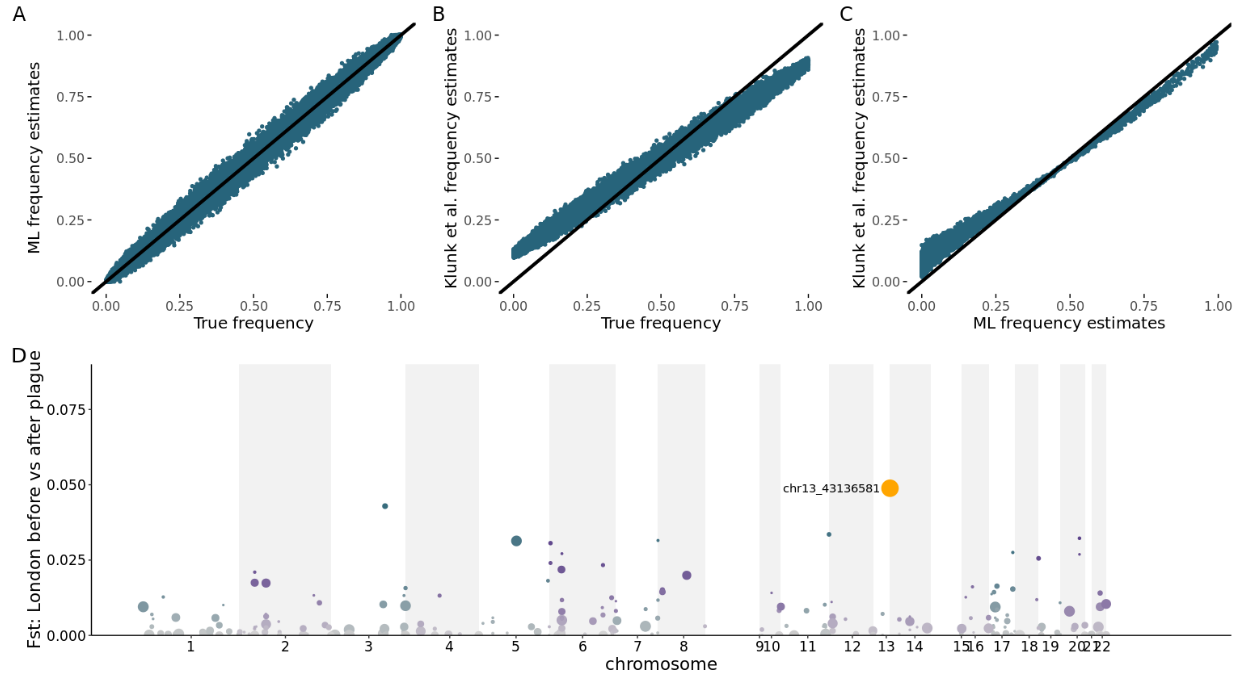
09:00 - 10:15 Lecture 3: Estimating allele frequencies, SNP calling and genotype calling from NGS data (Anders Albrechtsen)
10:30 - 12:00 Computer exercises III
12:00 - 01:00 Lunch (on your own)
01:00 - 02:15 Lecture 4: Imputation (Shyam Gopalakrishnan)
02:30 - 04:00 Computer exercises IV
04:15 - 05:00 Research talk: A genomic history of the Rapanui By Víctor Moreno Mayar
Canal boat trip
Wednesday - Population structure
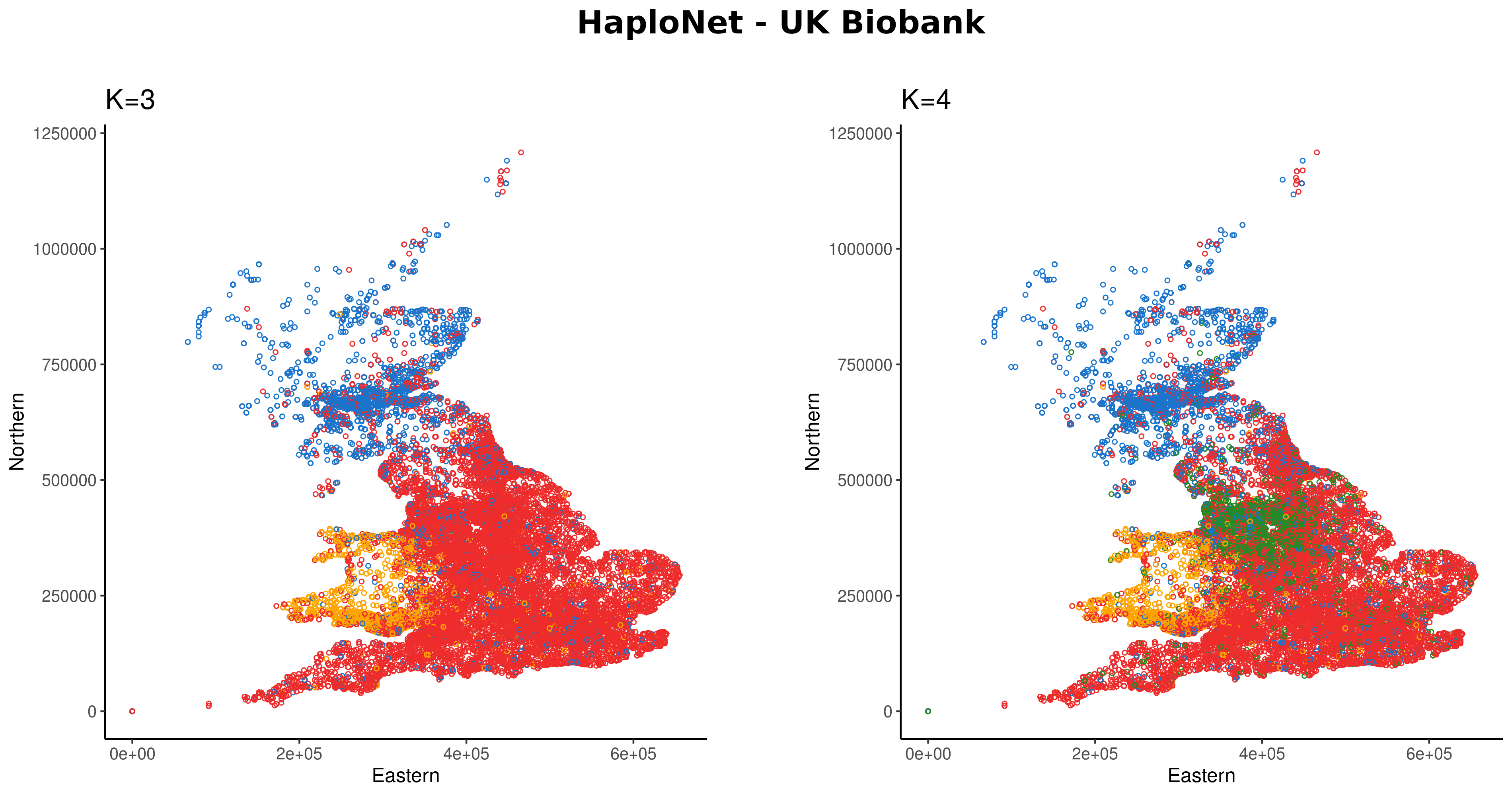
09:00 - 10:15 Lecture 5: Population structure and admixture (Ida Moltke)
10:30 - 12:00 Computer exercises V
12:00 - 01:00 Lunch (on your own)
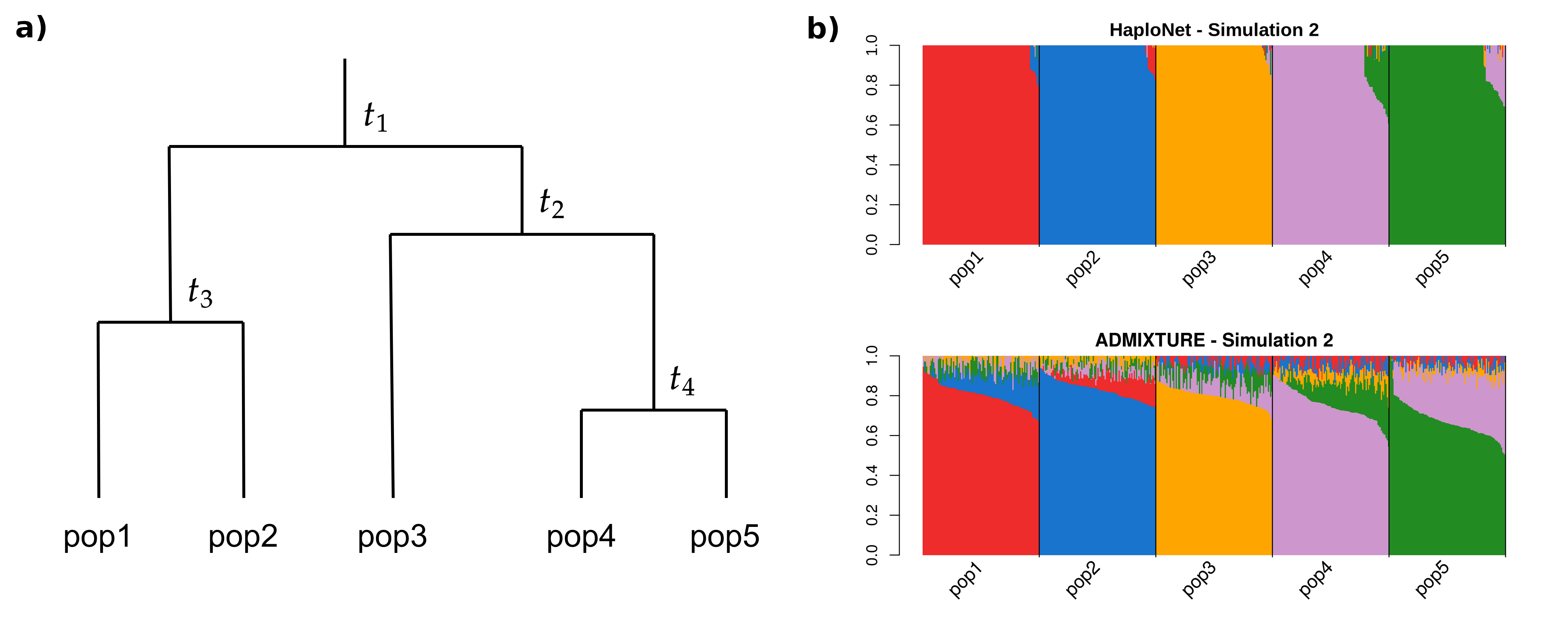
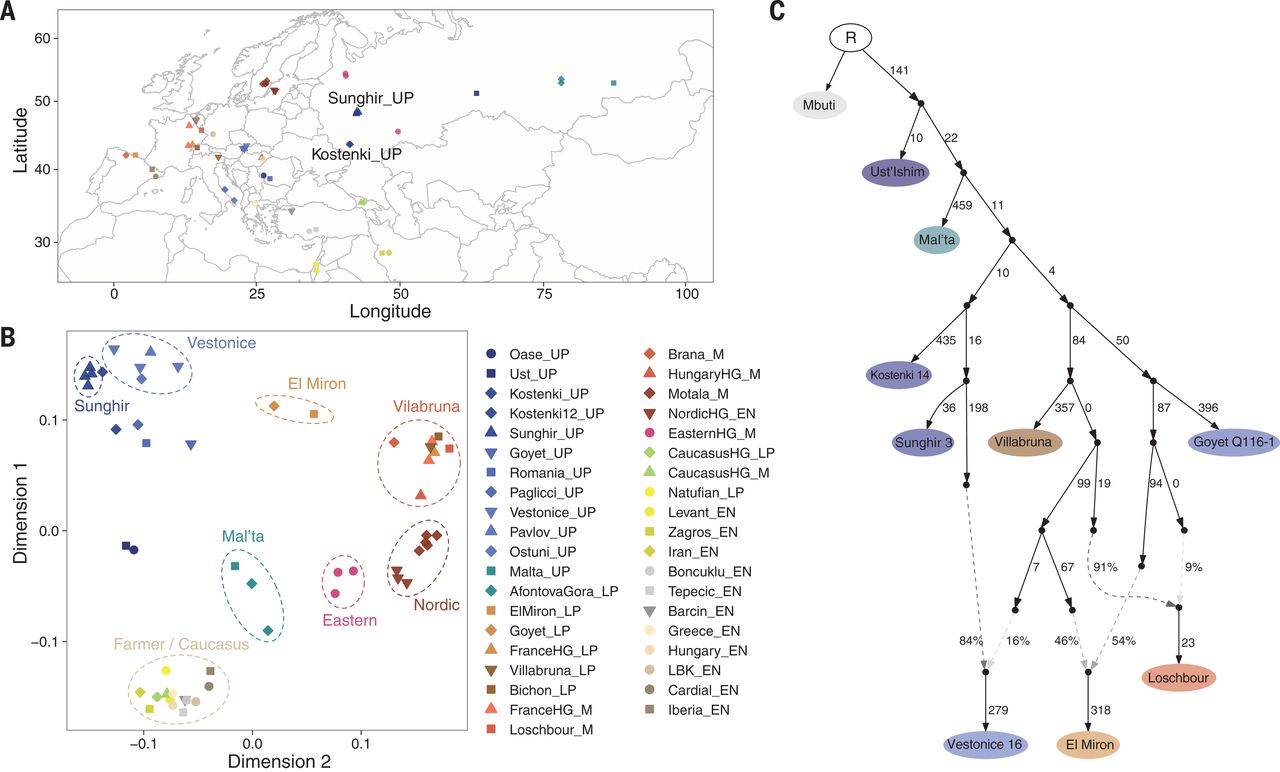
01:00 - 02:15 Lecture 6: D/f statistics and ancient geneflow (Martin Sikora)
02:30 - 04:00 Computer exercises VI
04:15 - 05:00 Research Talk: Ancient Pathogen Evolution and Epidemiology at Regional and Continental Scales by Martin Sikora
Thursday – Local ancestry inference and selection
09:00 - 10:15 Lecture 7: Local ancestry inference (Katia Bougiouri)
10:30 - 12:00 Computer exercises VII
12:00 - 01:00 Lunch (on your own)
01:00 - 02:15 Lecture 8: Detecting genomic regions under (positive) selection (Jazmin Ramos Madrigal)
02:30 - 04:00 Computer exercises VIII
04:15 - 05:00 Research Talk: Tracing the evolutionary history of the CCR5-delta32 deletion via ancient and modern genomes by Kirstine Ravn
Friday - PCA and demography
09:00 - 10:15 Lecture 9: PCA and genome masking (Anders Albrecthsen)
10:30 - 12:00 Computer exercises IX
12:00 - 01:00 Lunch (on your own)
01:00 - 02:15 Lecture 10: Demography Inference (Shyam Gopalakrishnan)
02:30 - 04:00 Computer exercises X
04:15 - 05:00 Research Talk: Vacuuming animal DNA from thin air by Kristine Bohmann
05:00 - ? Farewell drinks
Evaluation
Participants who have participated actively in all parts of the course and completed all exercises satisfactorily will be awarded a certificate of completion at the end of the course. The work load corresponds to 2.5 ECTS points which will be given for on site participant. Note that this workload includes one week of preparation. Reading material for this is available in the above course program.